Enzyme Function and Regulation
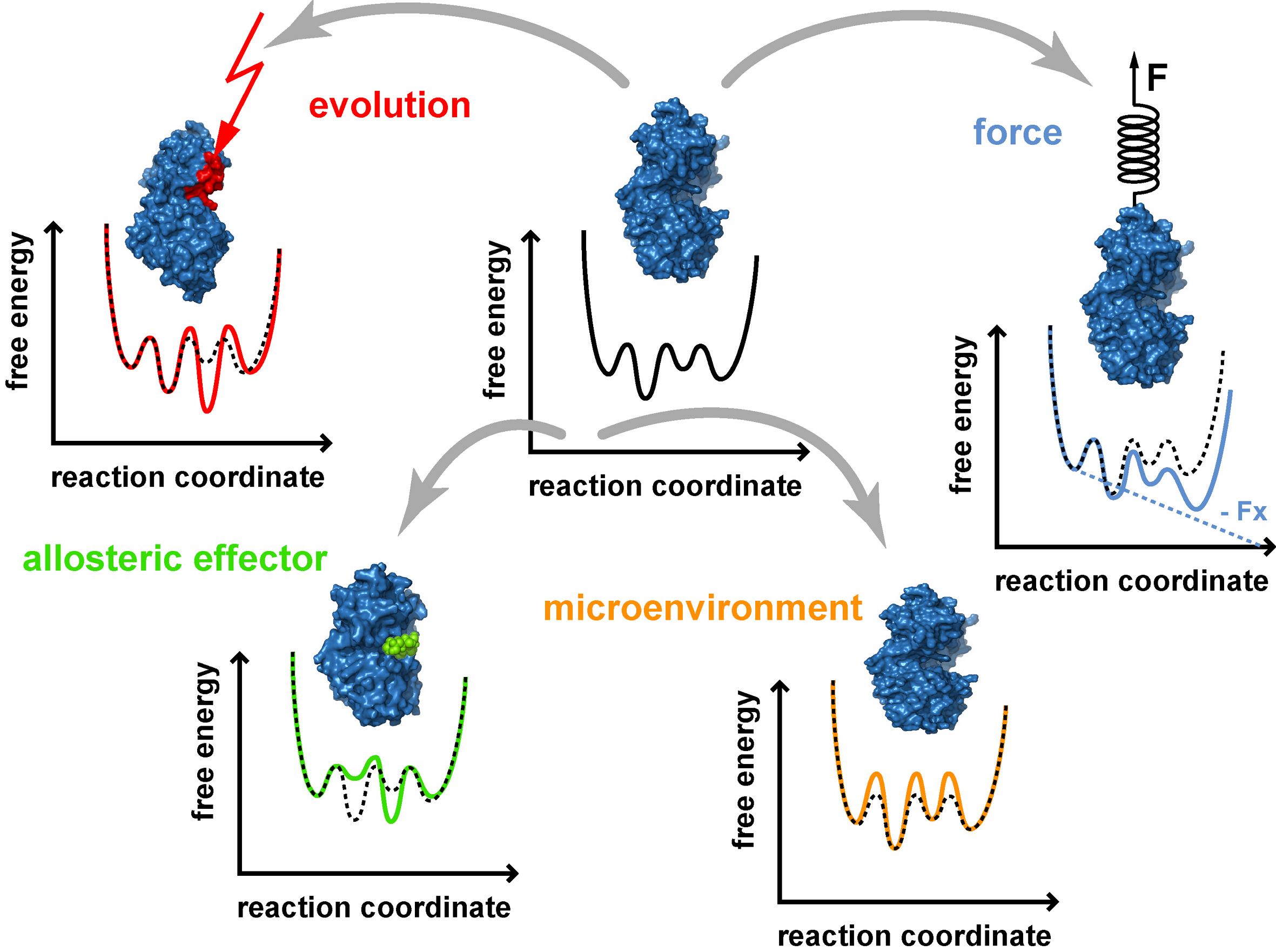
Single molecule experiments with lipase B from Candida antarctica (CalB) have shown that this enzyme has large fluctuations in the turnover rate and more importantly that it is only active for 3 % of the overall measurement time. Our current experiments aim at finding ways to increase the overall activity of CalB (with mutations, covalent modifications, buffer conditions, etc.) and to analyze the effect of these modifications at the level of individual enzymes.
We use a fluorescence based approach to detect enzymatic activity. Using a so-called fluorogenic substrate, which is cleaved by to enzyme to yield a fluorescent product molecule, we are able to detect individual enzymatic turnover reactions with a confocal fluorescence microscope. Surface immobilization of the enzyme ensures that the same enzyme can be monitored for extended periods of time. Subsequent data analysis of the times between individual turnover reactions ultimately yields the information how a specific modification alters the energy landscape and how the energy landscape determines the catalytic activity.
Bachelor and Master projects available
Collaborations:
Alan Rowan
Hans Engelkamp, Peter Christianen, Jan C. Maan
Jan van Hest
Johan Hofkens
Chun Biu Li
Klaus Schulten
Related publications:
- Blank, K., G. De Cremer, and J. Hofkens. (2009). Fluorescence-based analysis of enzymes at the single-molecule level. Biotechnol J 4:465-479.
- Engelkamp, H., N. S. Hatzakis, J. Hofkens, F. C. De Schryver, R. J. Nolte, and A. E. Rowan. (2006). Do enzymes sleep and work? ChemCommun (Camb):935-940.
- Flomenbom, O., K. Velonia, D. Loos, S. Masuo, M. Cotlet, Y. Engelborghs, J. Hofkens, A. E. Rowan, R. J. Nolte, M. Van der Auweraer, F. C. de Schryver, and J. Klafter. (2005). Stretched exponential decay and correlations in the catalytic activity of fluctuating single lipase molecules. Proc Natl Acad Sci USA 102:2368-2372.
- Velonia, K., O. Flomenbom, D. Loos, S. Masuo, M. Cotlet, Y. Engelborghs, J. Hofkens, A. E. Rowan, J. Klafter, R. J. Nolte, and F. C. de Schryver. (2005). Single-enzyme kinetics of CALB-catalyzed hydrolysis. Angew Chem Int Ed Engl 44:560-564.
Back to the top - back to index |